2019-
* corresponding author; # undergraduate student mentored; § graduate student mentored.
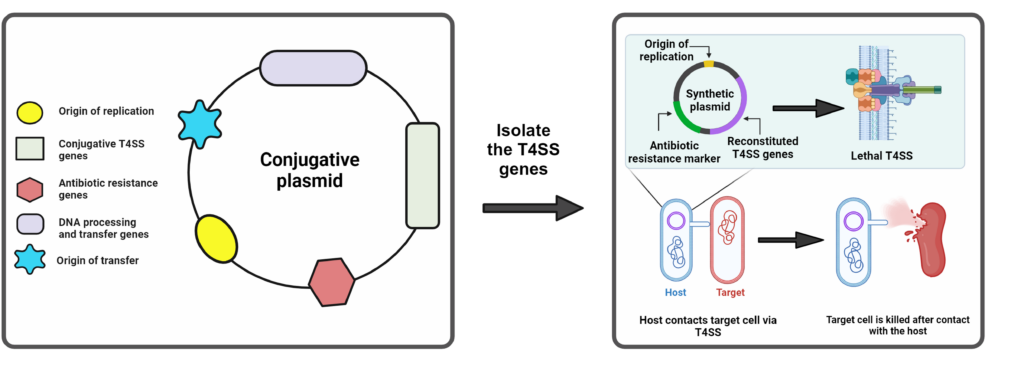
Gordils-Valentin, L.§, Ouyang, H.§, Qian, L.§, Hong, J.#, Zhu, X.*, Conjugative type IV secretion systems enable bacterial antagonism that operates independently of plasmid transfer. Commun. Biol., 2024, 7, 499.
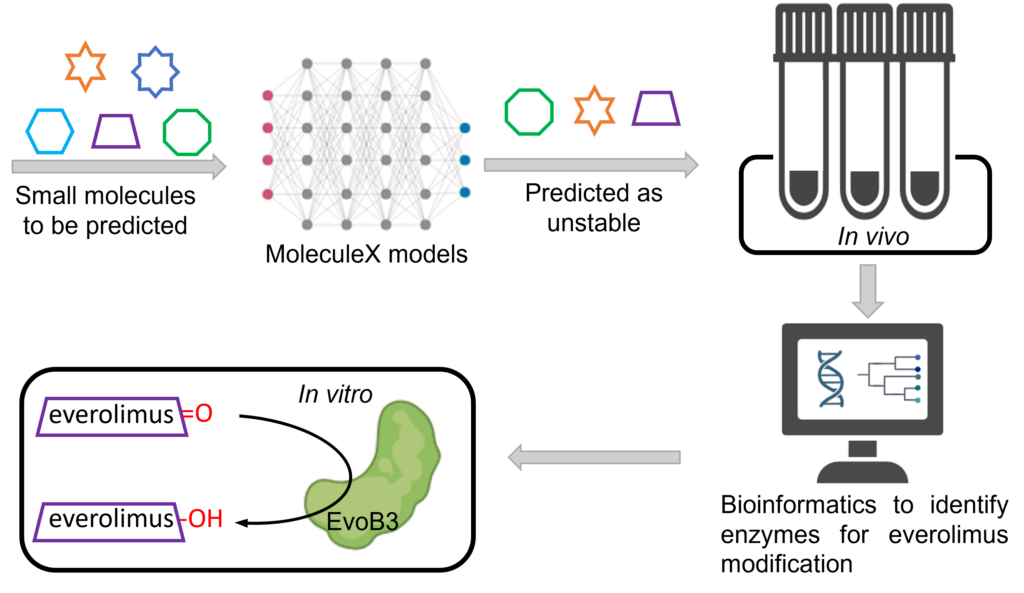
Ouyang, H.§, Xu, Z., Hong, J.#, Malroy, J.#, Qian, L.§, Ji, S., Zhu, X.*, “Mining the Metabolic Capacity of Clostridium sporogenes Aided by Machine Learning”, Angew. Chem. Int. Ed., 2024, 63, e202319925.
Qian, L.§, Mohanty, P., Jayaraman, A., Mittal, J., Zhu, X.*, “Specific residues and conformational plasticity define the substrate specificity of short-chain dehydrogenases/reductases”, J. Biol. Chem. 2024., 300(1), 105596.
Tao, X., Ouyang, H.§, Zhou, A.*, Wang, D., Matlock, H., Morgan, J., Ren, A., Mu, D., Pan., C., Zhu, X., Han, A., Zhou J.*, “Polyethylene Degradation by a Rhodococcous Strain Isolated from Naturally Weathered Plastic Waste Enrichment” Environ. Sci. Technol., 2023, 57, 37, 13901–13911

Qian, L.§, Ouyang, H.§, Gordils-Valentin, L.§, Hong, J.#, Jayaraman, A.*, Zhu, X.* “Identification of Gut Bacterial Enzymes for Keto-Reductive Metabolism of Xenobiotics” ACS Chem. Biol., 2022, 17(7), 1665–1671.

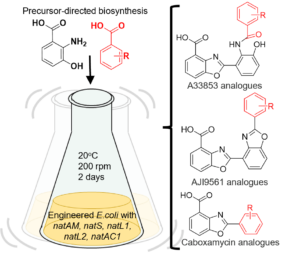
Ouyang, H.§, Hong, J.#, Malroy, J.#, Zhu, X.* “An E. coli-Based Biosynthetic Platform Expands the Structural Diversity of Natural Benzoxazoles” ACS Synth. Biol., 2021, 10(9), 2151-2158.
Li, J., Barber, C., Herman, N., Cai, W., Zafrir, E., Du, Y., Zhu, X., Skyrud, W., Zhang, W*. “Investigation of secondary metabolism in the industrial butanol hyper-producer Clostridium saccharoperbutylacetonicum N1-4.” J. Ind. Microbiol. & Biot. 2020, 47, 319–328.
Zhang, B., Rajakovich, L., Van Cura, D., Blaesi, E., Mitchell, A., Tysoe, C., Zhu, X., Streit, B., Rui, Z., Zhang, W.*, Boal, A.*, Krebs, C.*, Bollinger, J. M.* “Substrate-Triggered Formation of a Peroxo-Fe2(III/III) Intermediate during Fatty Acid Decarboxylation by UndA” J. Am. Chem. Soc. 2019,141(37), 14510-14514.
Before TAMU (2013-2019)
Seidel, J., Miao, P., Porterfield, W., Cai, W., Zhu, X., Kim, S., Hu, F., Bhattarai-Kline, S., Min, W., Zhang, W. “Structure–activity–distribution relationship study of anti-cancer antimycin-type depsipeptides” Chem. Commun. 2019, 55, 9379-9382.
Su, M., Zhu, X., Zhang, W. “Probing the Acyl Carrier Protein-Enzyme Interactions Within Terminal Alkyne Biosynthetic Machinery” AIChE J. 2018, 64, 4255-4261. (tribute to founders: Jay Bailey)
Zhu, X., Zhang, W. “Terminal Alkyne Biosynthesis in Marine Microbes” Methods Enzymol. 2018, 604, 89-112.
Harris, N. C., Sato, M., Herman, N. A., Twigg, F., Cai, W., Liu, J., Zhu, X., Downey, J., Khalaf, R., Martin, J., Koshino, H., Zhang, W. “Biosynthesis of isonitrile lipopeptides by conserved non-ribosomal peptide synthetase gene clusters in Actinobacteria” Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 7025-7030.
Zhu, X., Shieh, P., Su, M., Bertozzi, C. R., Zhang, W. “A fluorogenic screening platform enables directed evolution of an alkyne biosynthetic tool.” Chem. Commun. 2016, 52, 11239-11242.
Liu, J., Zhu, X., Kim, S., Zhang, W. “Antimycin-type depsipeptides: discovery, biosynthesis, chemical synthesis, and bioactivities.” Nat. Prod. Rep. 2016, 33, 1146-1165.
Rui, Z., Harris, N. C., Zhu, X., Huang, W., Zhang, W. “Discovery of a family of desaturase-like enzymes for 1-alkene biosynthesis.” ACS Catal. 2015, 5, 7091-7094.
Liu, J., Zhu, X., Zhang, W. “Identifying the minimal enzymes required for biosynthesis of epoxyketone proteasome inhibitors.” ChemBioChem. 2015, 16, 2585-2589.
Zhu, X., Su, M., Manickam, K., Zhang, W. “Bacterial genome mining of enzymatic tools for alkyne biosynthesis.” ACS Chem. Biol. 2015, 10, 2785-2793.
Zhu, X., Zhang, W. “Tagging polyketides/non-ribosomal peptides with a clickable functionality and applications.” Front. Chem. 2015, 3: 11.
Zhu, X., Liu, J., Zhang, W. “De novo biosynthesis of terminal alkyne-labeled natural products.” Nat. Chem. Biol. 2015, 11, 115-120. (selected as NCB’s greatest hit) news & views NCB’s greatest hits
Liu, J., Zhu, X., Seipke, R. F., Zhang, W. “Biosynthesis of antimycins with a reconstituted 3-formamidosalicylate pharmacophore in Escherichia coli.” ACS Synth. Biol. 2015, 4, 559-565.
Rui, Z., Li, X., Zhu, X., Liu, J., Domigan, B., Barr, I., Cate, J., Zhang, W. “Microbial biosynthesis of medium-chain 1-alkenes by a non-heme iron oxidase.” Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 18237-18242. highlight in Angew. Chem.
Sandy, M., Zhu, X., Rui, Z., Zhang, W. “Characterization of AntB, a promiscuous acyltransferase involved in antimycin biosynthesis.” Org. Lett. 2013, 15, 3396-3399.